High-Performance Computing in Action – Sano Researchers at KU KDM 2025
The Seventeenth HPC Users’ Conference (KUKDM 2025) was held in Zakopane from April 2–4, bringing together the scientific and technological communities in an interdisciplinary setting.
This year’s edition introduced an updated formula, offering enhanced opportunities for scientific training and direct consultations with the PLGrid support team. The program featured the latest developments in high-performance and quantum computing, and showcased research enabled by Cyfronet’s infrastructure.
The conference focused on showcasing Cyfronet’s infrastructure, including its computing power, software tools, and user services. It offered insights into emerging trends in computing and networking—such as quantum computing capabilities—and featured case studies of research conducted using Cyfronet’s resources. Participants also had the chance to engage with technology providers and take part in hands-on training, expert consultations, and open discussions with Cyfronet staff.
Sano Centre for Computational Medicine at KU KDM 2025
Sano Centre for Computational Medicine was actively represented at the event by our researchers, who presented several posters highlighting the application of HPC technologies in computational medicine.
Posters presented by our team included:
Dominika Ciupek: “Bipolar disorder characterization from diffusion MRI: A deep learning approach”
Co-authors: Maciej Malawski, Tomasz Pięciak.
Taras Zhyhulin: “Toolkit for Managing Research Data in Medical Simulations on HPC Infrastructure”
Co-authors: Karol Zając, Piotr Nowakowski, Maciej Malawski, Jan Meizner, Marek Kasztelnik, Piotr Połeć.
“Medical simulations are typically computationally and memory-intensive, requiring robust data management strategies and infrastructure to enable internal sharing and transfer of data to HPC systems. The results of such simulations, input datasets, and related research data should be published to support the broader scientific community and uphold Open Science principles. Furthermore, researchers should be incentivized to contribute to the datasets they use to help expand and improve them.
The toolkit presented in the poster facilitates these practices and encourages their adoption among scientists. It comprises several components supporting different research stages. To improve data management during simulations, the Model Execution Environment (MEE) platform—designed by Cyfronet and developed with Sano—has been integrated with Dataverse, Zenodo, and InvenioRDM repositories. This integration enables seamless automatic data transfers between repositories and HPC resources, eliminating the need for intermediate storage.
An equally important aspect is internal team sharing and publishing data at the research’s conclusion. To promote Open Science and FAIR principles, Sano Dataverse was deployed and configured for our research teams. It allows users to publish data with DOIs via DataCite. To further increase reach and credibility, Sano Dataverse is integrated into RODBUK, the Kraków Open Research Data Repository—a Polish federation of Dataverse instances. RODBUK aggregates members’ data on its main page, improving discoverability and citation potential, while its robust access controls and review policies enhance trustworthiness.
We also propose a rule-based data sharing strategy to encourage contributions to valuable datasets. This involves publishing data under restrictions or embargoes and allowing users to request access by providing a reason and a sample contribution. If the dataset owner approves, the user is granted early access and can add their data. This mechanism fosters collaboration and helps datasets grow in value and insight potential.
Overall, the toolkit enhances research efficiency and promotes Open Science by streamlining data sharing and encouraging good practices. It supports both Sano researchers and the broader scientific community by providing tools and strategies for sustainable, collaborative research data management.”Taras Zhyhulin
Kamil Burkiewicz: “Serverless Transcriptomics on the HPC cluster”
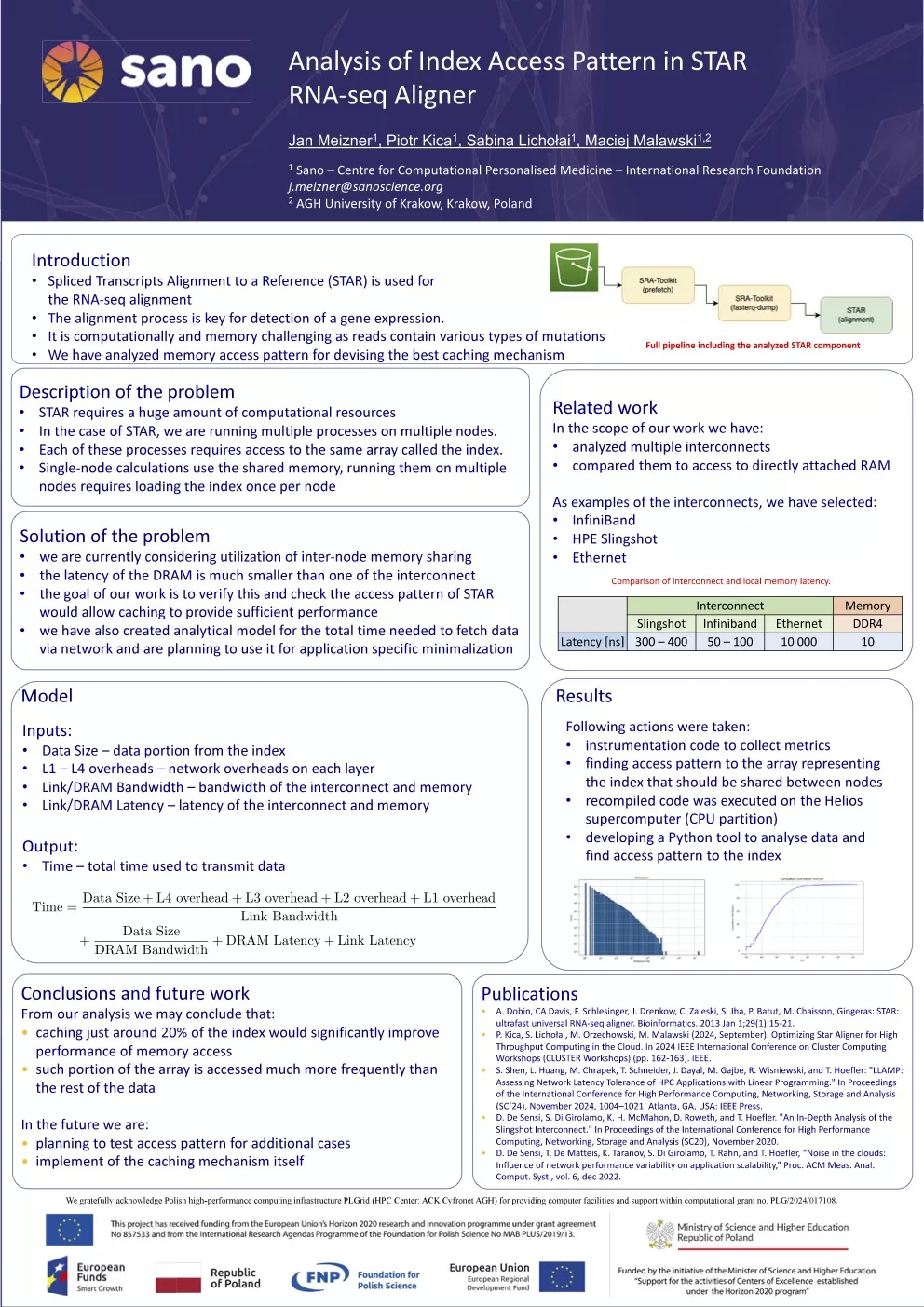
Jan Meizner: “Analysis of Index Access Pattern in STAR RNA-seq Aligner”
Co-authors: Piotr Kica, Sabina Lichołai, Maciej Malawski.
Karol Zając: “Verification, Validation and Uncertainty Quantification Workflows on High Performance Computers”
“Karol Zając presented a poster on the acceleration of Verification, Validation, and Uncertainty Quantification (VVUQ) workflows using High-Performance Computing (HPC) resources. The work showcased how the EasyVVUQ framework was employed to manage UQ and sensitivity analysis experiments, while Dask (via Dask JobQueue and Dask-MPI) was used independently to parallelize and scale the execution of simulation tasks. This setup enabled efficient distribution of workloads across HPC nodes, significantly reducing computational time and improving throughput. The approach was validated on the Ares cluster at Cyfronet and provides a robust foundation for executing complex VVUQ studies involving both Python-based and external MPI applications.”
Karol Zając
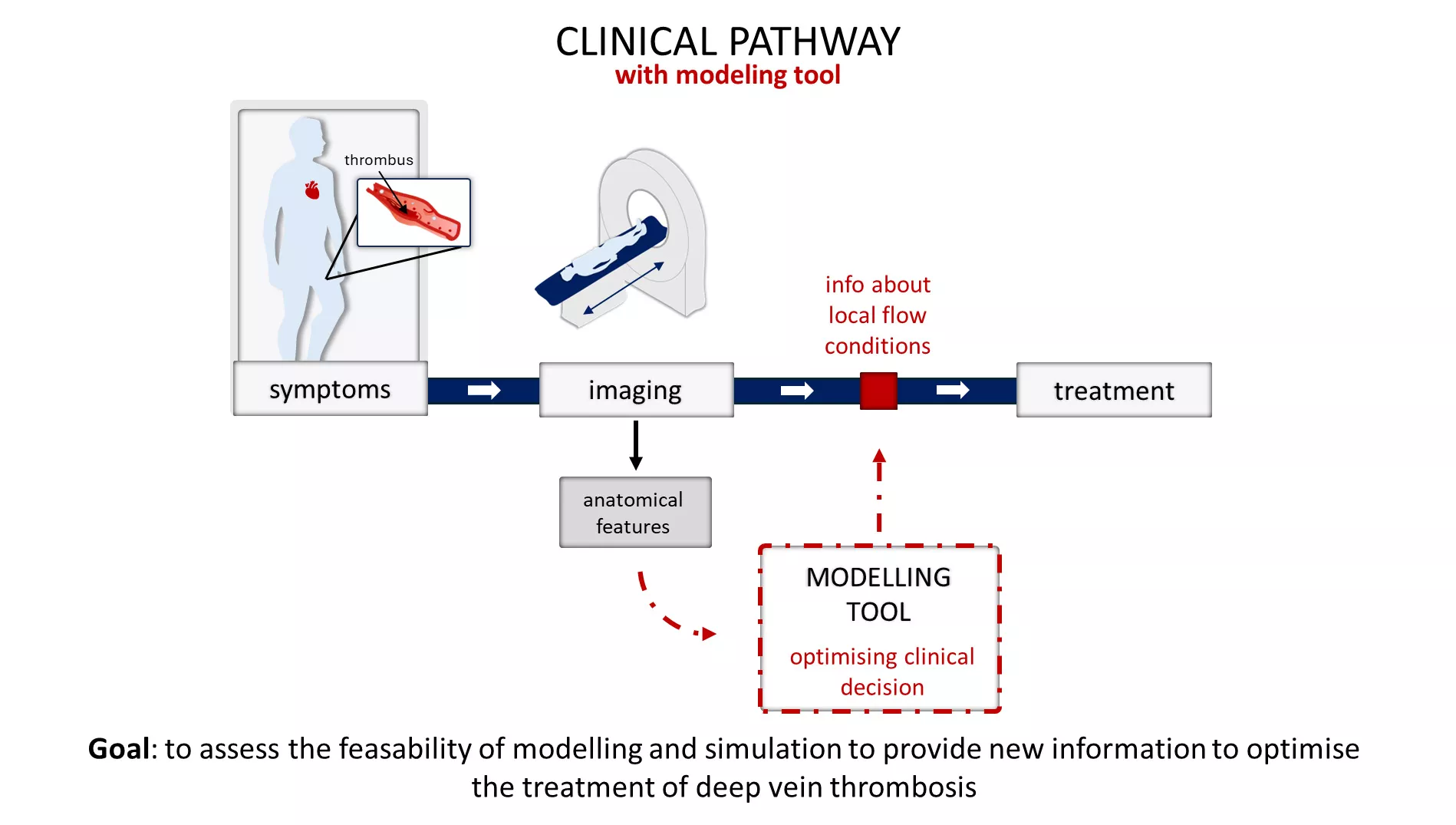
Karolina Tlałka: “Using High Performance Computing for the Quantification and Personalization of Cardiovascular Models”
Co-authors: Harry Saxton, Ian Halliday, Andrew Narracott, Maciej Malawski.
The work titled Using High Performance Computing for the Quantification and Personalization of Cardiovascular Models presents how high performance computing (HPC) can be applied to analyze the convergence of a global optimizer used in the personalization of a regulated cardiovascular model. Leveraging HPC reduced computation time by a factor of 31 — a significant performance improvement compared to running the model on a local machine.
“The KUKDM 2025 conference was a great opportunity to explore how HPC is utilized across various scientific disciplines. We also participated in insightful and hands-on workshops. The knowledge gained from the conference will support more efficient use of HPC resources.”
Karolina Tlałka
These contributions reflect the diversity and depth of our research efforts at Sano and underline the importance of HPC in advancing computational approaches to medical challenges.