Sano Researchers Present Work at ICCS 2025 in Singapore
Advancing interdisciplinary science through computational innovation, Sano – Centre for Computational Medicine joined researchers from around the globe at ICCS 2025 in Singapore, held from 7 to 9 July.
The event convened leading experts from multiple disciplines to explore the latest developments in computational approaches to complex scientific problems. The event brought together scientists working at the intersection of mathematics, computer science, and applied domains such as life sciences, physics, and engineering. ICCS has long been recognized as a premier platform for advancing interdisciplinary discussions and presenting state-of-the-art computational methods.
The theme of this year’s conference – “Making Complex Systems Tractable Through Computational Science” – highlighted the growing importance of scalable algorithms, and high-performance data analytics in addressing modern scientific challenges.
Sano researchers contributed a diverse range of presentations and posters:
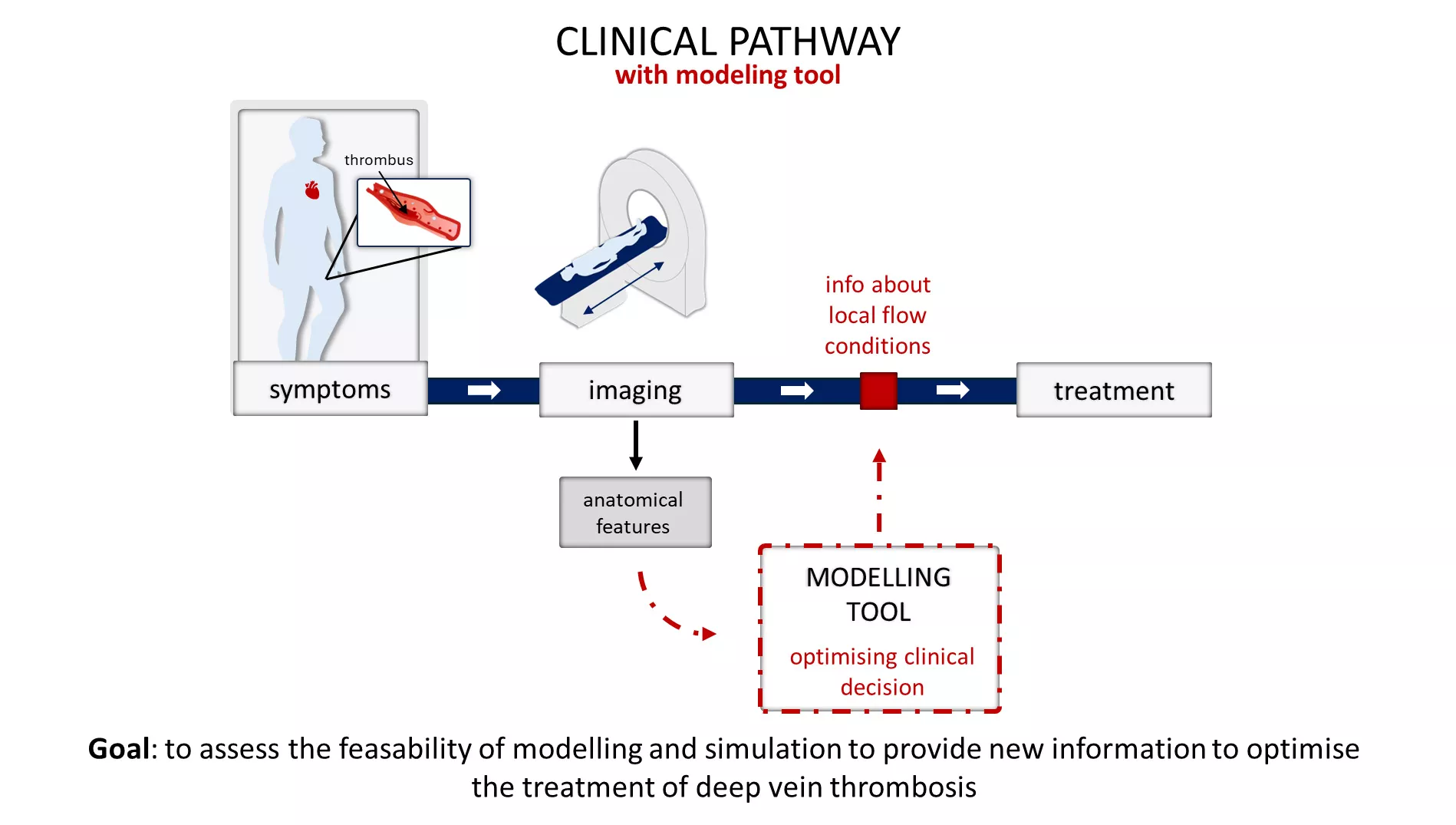
Magdalena Otta, PhD student in the Extreme-scale Data and Computing team, gave a talk titled “Towards sensitivity analysis: 3D venous modelling in the lower limb,” presenting computational models supporting vascular research. The work is based on a published paper that reported an analysis of a set of idealised 3D geometries of iliac vein unification to assess the importance of shape variability, inflow conditions, and viscosity on local haemodynamics – specifically on the wall shear stress metrics. A detailed steady state analysis focused on the wall shear stress distributions below three thresholds (<0.15 Pa, <0.10 Pa <0.05 [Pa]), and transient state focused on the oscillating shear index above three thresholds (>0.25, >0.35, >0.45). All the variations implemented had an effect on the size and shape of the absolute vein wall area subject to the shear metrics of choice under the assumed flow conditions. The results will serve as a basis for the interpretation of patient-specific geometries of the iliac vein unification affected by deep vein thrombosis.


Jakub Klimczak delivered a presentation titled “Explainable Artificial Intelligence for Bioactivity Prediction: Unveiling the Challenges with Curated CDK2/4/6 Breast Cancer Dataset,” focusing on the transparency and interpretability of AI models in pharmacologica applications. The talk addressed key limitations of traditional explainability approaches that rely on assumptions of feature independence and additive contributions, which often fail to capture the complexity of chemical data. Jakub showcased analyses of fingerprint-based and molecular graph models, highlighting their strong predictive performance but limited actionable insights for medicinal chemists. To bridge this gap, he proposed a novel benchmark grounded in the pharmacophore concept and introduced PharmacoScore, a new metric designed to evaluate whether machine learning explanations correctly prioritize critical pharmacophore elements over irrelevant features.
Piotr Kica presented a poster titled “Accelerating Cloud-Based Transcriptomics: Performance Analysis and Optimization of the STAR Aligner Workflow,” co-authored with Sabina Lichołai, Michał Orzechowski, and Maciej Malawski. The work is based on a published study that proposes a scalable, cloud-native architecture for executing STAR — one of the most widely used RNA-sequence aligners — on large-scale transcriptomic datasets. The team implemented a series of optimization techniques to significantly reduce execution time and cloud computing costs. A key improvement, the early stopping mechanism, enabled a 23% reduction in total alignment time. The study also identifies optimal EC2 instance types and evaluates the use of spot instances for cost-efficient computing. The methodology was validated through medium- and large-scale experiments.


Wieslaw L. Nowinski, Senior Researcher at Sano, gave a talk titled “From the synaptome to the connectome: data bigness estimation for the human connectome at the nanoscale.” Drawing on his publication, Nowinski introduced a set of wireframe and geometric models for estimating the size and data requirements of the human nanoscale connectome. His presentation highlighted six variations of the connectomic model and 36 estimation cases, demonstrating that the storage demands for fully representing the human brain’s nanoscale wiring—from synapses to entire circuits—exceed even the capacities of today’s most advanced supercomputers. This pioneering work marks the first attempt to provide precise estimations of the data needed to model the entire human connectome at nanoscale resolution.
Paweł Szczerbiak, postdoctoral researcher in the Structural and Functional Genomics Group, and Maciej Malawski, research team leader of Extreme-scale Data and Computing, also represented Sano at the event. They actively engaged in sessions on computational genomics, cloud architecture, quantum machine learning, and scalable infrastructures. Notably, the event facilitated valuable networking opportunities, laying the groundwork for potential future collaborations.

Participation in ICCS 2025 underscores Sano’s ongoing commitment to advancing computational tools for precision medicine and contributing to the global scientific community working at the forefront of computational science and high-performance biomedical data processing.
While in Singapore for ICCS 2025, the Sano team also took the opportunity to visit the NUS Mind Science Centre (NMSC). This pioneering institution, affiliated with the National University of Singapore, promotes an interdisciplinary approach to understanding mental resilience, ageing, and psychological well-being. Its research and outreach programmes bridge neuroscience, behavioural science, and preventive healthcare, with a strong emphasis on real-world applications in education, eldercare, and youth mental health. By combining academic rigor with community engagement, NMSC stands at the forefront of efforts to translate brain science into impactful, evidence-based interventions across the human lifespan.