Sano researchers co-create AI-powered platform to explore protein structures
Scientists from Sano Centre for Computational Medicine have co-authored research that combines artificial intelligence with protein biology to accelerate discoveries in medicine. The work, published in Nature Communications, presents a new interactive online platform that organizes the world’s largest datasets of protein structures and makes them accessible to the global scientific community.
A tool for exploring the landscape of proteins
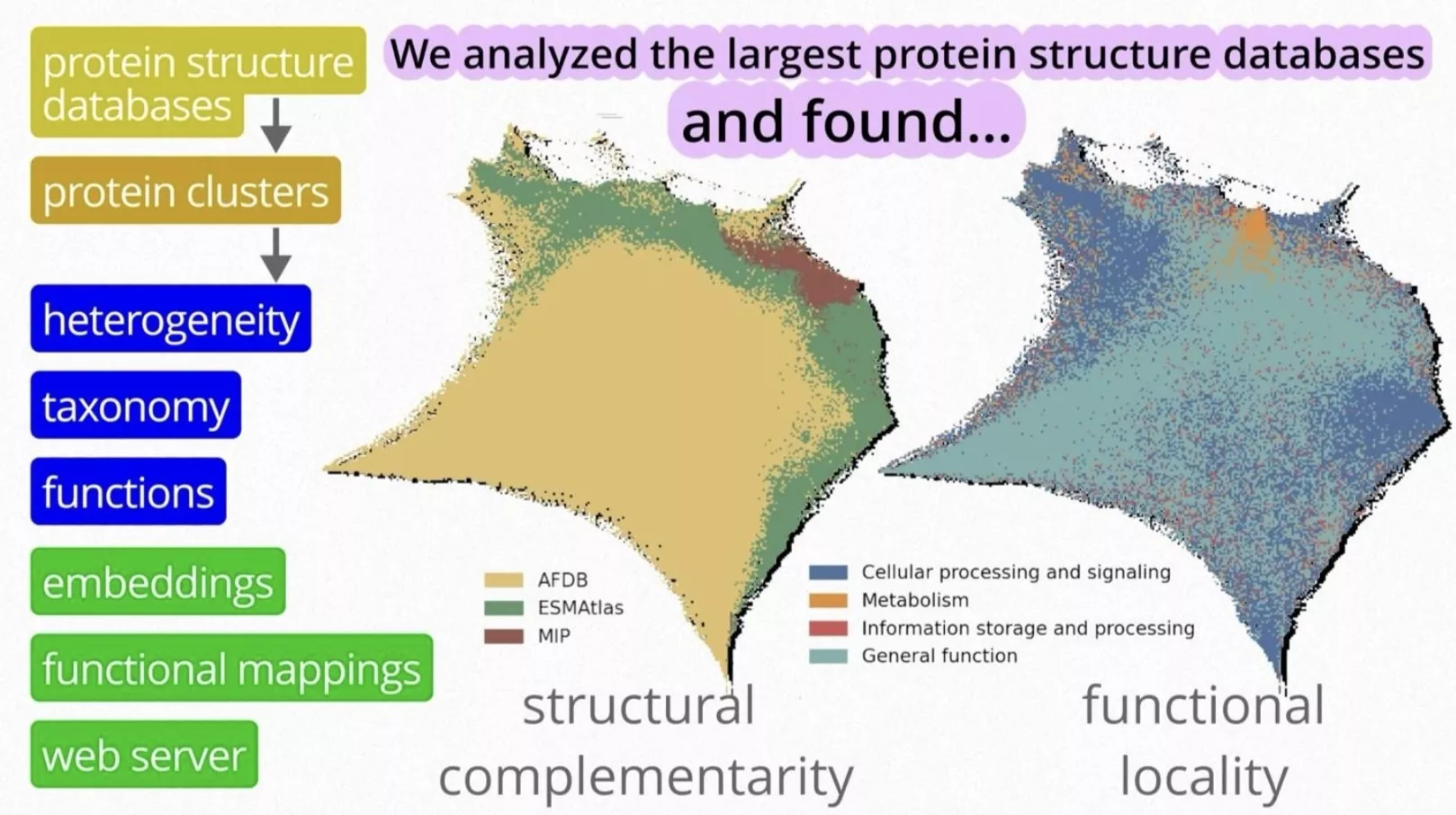
The new platform – available at protein-structure-landscape.sano.science – enables researchers to visually explore and analyze nearly one billion predicted protein structures. By integrating data from three of the largest resources – AlphaFold (awarded the 2024 Nobel Prize in Chemistry), ESMAtlas, and the Microbiome Immunity Project – the tool provides a unified “map” of protein space.
“Our goal was not only to organize these massive datasets but also to create a resource that helps us better understand protein biology,” explains Dr. Tomasz Kościołek, co-author of the study and leader of the Structural and Functional Genomics team at Sano.
The team demonstrated that proteins with similar biological functions often cluster together in the data landscape – a phenomenon they call “functional locality.”
AI methods reveal hidden protein functions
In addition to mapping the known data, the researchers applied advanced AI methods such as deepFRI, which can predict protein functions even when their sequences differ from previously characterized examples. This approach allowed the discovery of previously unknown variants, including proteins potentially involved in lipid transport or biochemical processes in extreme environments.
“We believe our platform will contribute to the development of better diagnostic and therapeutic methods, while also helping us understand molecular life at the structural level,” says Dr. Paweł Szczerbiak, first author of the study and researcher at Sano.
An international collaboration led from Krakow
The project was developed in collaboration with Jagiellonian University, the Flatiron Institute (USA), and the Open Molecular Software Foundation (USA).
Global impact and open access
The platform is freely available to scientists around the world. It can be used for microbiome research, studies on protein evolution, and in the design of new drugs and therapies. Its intuitive, visual interface also makes it a valuable educational tool for students and young researchers.
⸻
🔗 Read the full publication in Nature Communications: https://www.nature.com/articles/s41467-025-63250-3
🔗 Explore the interactive protein map: protein-structure-landscape.sano.science