
Krakow Conference on Computational Medicine 2025
Enhancing Virtual Human Twin with AI solutions
October 15–17, 2025, KrakowTwo films about the Krakow Conference on Computational Medicine
Realisation: Jacek Przybylski
Filming, sound, editing: Marian Przybylski
Support: Marian Bubak and Maria Sendecka – Sano
Production: ACC Cyfronet AGH
Proceedings KCCM 2025
Organisers
- Sano Centre for Computational Medicine, Krakow Poland
- Faculty of Computer Science AGH Krakow, Poland
- Academic Computer Center AGH Krakow, Poland
Scope and topics
The conference’s motto is “Enhancing Virtual Human Twin with AI solutions“. Personalized medicine, focusing on the development of in-silico methods replacing in-vivo and in-vitro methods, should more effectively use the solutions brought by the AI revolution based on machine learning and data analysis methods; perceived not as competitive, but as supporting existing modeling and simulation methods.
Given the Organisers’ expertise in both computer simulation and artificial intelligence, the Conference will be an excellent opportunity to gain greater interaction between the communities working in these two fields. Computer technologies and high-performance computing are of key importance for progress in computational medicine and therefore an additional advantage of the Conference will be the inclusion of technical aspects of the use of new computing infrastructures.
The organisationof the Conference is the result of the experience gained by the Sano team during Sano Science Day (2023, 2024) and cooperation in the Life Science Open Space organisation (since 2019) and as well as on a very broad experience of the Faculty of Computer Science AGH and Academic Computer Centre Cyfronet AGH in this area.
Conference topics include
- Ethical, legal, and social issues in VHT
- Mathematical medical models
- Multiscale modelling
- Computational modelling of organs and diseases
- Patient data management and processing
- Methods of acquisition, storage and retrieval of information in medicine
- Analysis of medical images
- Machine learning models for healthcare
- Computer simulations using advanced computing infrastructures
- Surgical planning tools
- Model and simulation reproducibility and credibility
- Clinical decision support systems based on artificial intelligence
- Towards the Virtual Human Twin platform
Keynote Lectures
Ewa Deelman – University of Southern California, Information Sciences Institute
Opportunities for AI in Modern Cyberinfrastructure: The Case of Scientific Workflow Management
Over the last two decades, scientific workflow management systems (WMSs) have enabled the execution of complex, multi-task applications on a variety of computational platforms, including today’s exascale systems. They ensure efficient execution of computational and data management tasks, adhering to their data and control dependencies. During workflow execution, WMSs monitor the execution of tasks, detect anomalies and failures, and deploy recovery mechanisms when needed. If the workflow cannot be successfully executed, the WMS provides debugging information to help the scientist or cyberinfrastructure (CI) operator fix the problem. However, as workflows and CI grow in scale, heterogeneity, and complexity, traditional WMS approaches face challenges in scalability, adaptability and resilience.
Although research in WMS has explored a number of avenues from workflow composition using semantic technologies to resource provisioning, workflow scheduling, fault-tolerance, and provenance tracking, the methods employed were often heuristics-based and limited in their applicability. With semantic technologies, there was a significant amount of investment in the generation of appropriate ontologies that needed to happen in order to ease workflow composition. Today, AI technologies are making a significant impact on every aspect of our lives, and they are being used in science as well. Although they are also applicable to CI, potentially making it more performant and resilient, they have not yet made significant inroads. In this work, we focus on the exploration of the use of AI in the case of workflow management systems, exploring its use throughout the scientific workflow lifecycle from workflow design to workflow execution. We investigate the use of AI in the context of the existing Pegasus WMS, exploring embedding AI in current systems as well as reimaging workflow management to be fully distributed and resilient. This new model is inspired by enhanced swarm intelligence (SI), designed to dynamically adapt to failures and optimize the overall system.
The Pegasus WMS. Pegasus pioneered the use of planning in WMSs, enabling users to focus on their science by describing their workflows in a resource-independent way. Pegasus takes that description and automatically maps the jobs onto heterogeneous resources, determines the necessary data transfers between jobs, and optimizes the workflow for performance and reliability. The result is an executable workflow that includes compute job submit scripts and data management jobs for the target CI. Pegasus has a notion of the submit host from where the system submits jobs to multiple distributed resources within the CI ecosystem: HTC (high-throughput computing) and high-performance computing (HPC) resources, campus clusters, user-provisioned clouds, and the edge. Pegasus workflows are easy to compose using Python, Java, and R APIs as well as in Jupyter Notebooks, and are portable across heterogeneous CI.
PegasusAI: Integrating AI throughout the workflow lifecycle. Whereas Pegasus uses simple heuristics, PegasusAI, a new project started in 2025, explores a variety of AI technologies throughout the entire workflow lifecycle. For workflow composition, we are exploring retrieval-augmented generation, combining the generative capabilities of large language models (LLMs) with retrieval mechanisms, to support workflow discovery and composition by providing context-specific suggestions that users can adapt as needed. Today, the user identifies or provisions the resources needed for execution. Once resources are provisioned, the WMS plans the workflow onto the resources using static heuristics. PegasusAI will fully automate the resource provisioning and planning steps. The planner will identify the types of resources needed, and then the provisioner will acquire the needed resources before the planner maps the workflow onto them. To make smart decisions, we explore a variety of AI models, including neural network-based models and probabilistic models, to learn about resource and job performance and robustness. Once the workflow is sent for execution, PegasusAI monitors the jobs in real time and performs anomaly and error detection (slow network, overloaded system, application error) using techniques such as Graph Neural Networks, LLMs, or autoencoders that we explored in prior work and decide whether to adapt the execution or the mapping or suggest to the user to modify the workflow. As results are being generated, PegasusAI learns about successful workflow design patterns and their components.
Part of the challenge of adopting AI for CI may be the quick pace of AI development as well as the challenges associated with the deployment of the technologies. Overtime, one needs to have a way of migrating between AI models as the new generations become more capable. This includes the evaluation of new technologies in terms of the validity of the solution, performance, and resource needs, as well as potential additional fine-tuning, “catching up” with the latest model to what the previous version already learned about the CI context. Issues of deployment, such as availability of appropriate computational and storage resources over time to support learning and inferencing, issues of resilience when models are embedded in the CI, or connectivity when queried remotely, need to be taken into account.
SWARM: Scientific Workflow Applications on Resilient Metasystem. Pegasus and PegasusAI are designed to be production-level capabilities. Pegasus is being used today in astronomy, bioinformatics, climate and weather modeling, ecology, earthquake science, gravitational-wave physics, materials science, chemistry, and AI/ML for science.
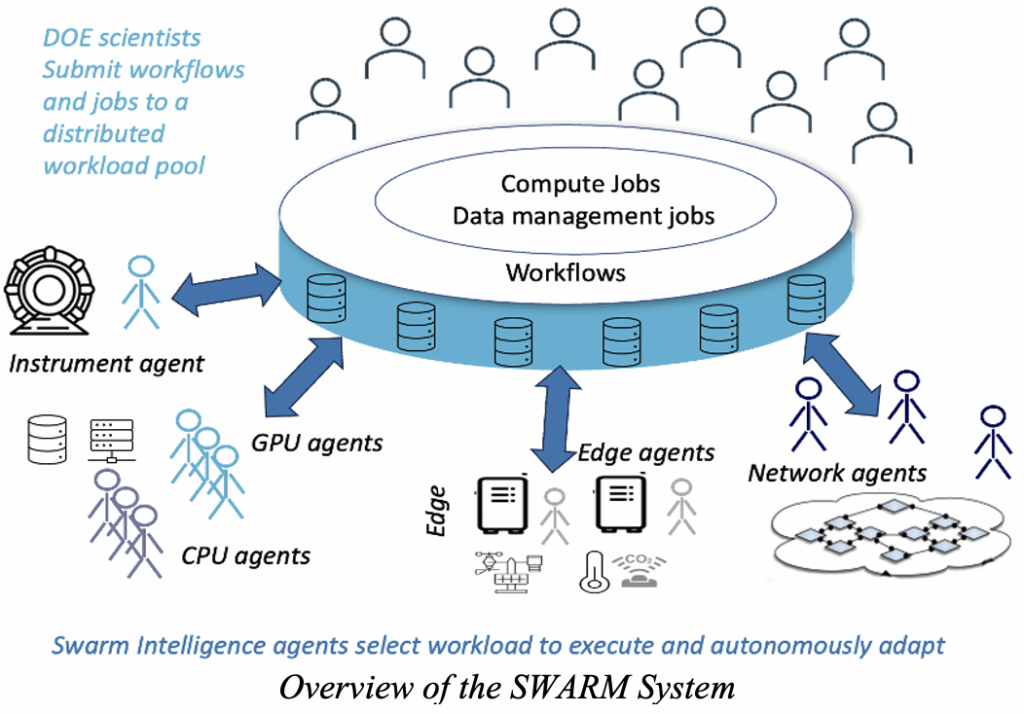
PegasusAI will build on the current capabilities and incrementally incorporate AI to make the system easier to use, more performant, and robust. However, there are limitations to the current approach, as the system is centralized. SWARM, on the other hand, reimagines the WMS as fully distributed and agentic, making agents responsible for the distributed compute resources across the computing continuum, from the edge to the core.
SWARM integrates swarm intelligence, consensus protocols, and optimized network overlays. Instead of relying on a centralized scheduler, SWARM models job scheduling, data management, and fault recovery as autonomous yet cooperative agents—ranging from lightweight edge agents to LLM-enhanced cognitive agents—that self-monitor, diagnose, and adapt to failures. Consensus plays a central role: heterogeneous agents achieve agreement on job selection and resource allocation using tailored consensus functions and resilient algorithms, such as Practical Byzantine Fault Tolerance and novel greedy variants, which ensure progress even under failures. To address scalability and efficiency, SWARM employs dynamic overlay networks, where the communication topology adapts to resource capabilities and network conditions, balancing local and global connectivity to minimize overhead while maintaining robustness. The multi-agent work includes a fault-tolerant, fully distributed scheduling architecture that eliminates the single points of failure common in centralized systems. Independent agents manage local resources, monitor system health, and coordinate through competitive bidding, achieving near-optimal job allocation without heavy consensus overhead. SWARN is meant to be a research effort that explores ideas of agentic AI for CI. However, we expect that some of the ideas will be valuable in production.
Acknowledgements: PegasusAI is supported by the U.S. National Science Foundation under grant #2513101. SWARM is funded by U.S. Department of Energy under grant #DE-SC0024387.
Ewa Deelman, University of Southern California
Liesbet Geris – KU Leuven, VPH Institute
Building the Virtual Human Twin: from an engaged ecosystem to an incipient infrastructure
Following the definition of the European Commission, a Virtual -Human Twin (VHT) is a digital representation of a human health or disease state. VHTs refer to different levels of human anatomy (e.g. cells, tissues, organs or organ systems). They are built using software models and data and are designed to mimic and predict behaviour of their physical counterparts, including interaction with additional diseases a person may have. The key potential in health and care of this technology is related to targeted prevention, tailored clinical pathways, and to supporting healthcare professionals in virtual environments. Examples include implementation of clinical trials for medicines and devices, medical training, surgical intervention planning, and several other potential use cases in virtual world environments. A public VHT infrastructure is the subject of an ongoing tender. This infrastructure should enable the pooling of resources and assets (data, models, algorithms, computing power, storage etc.) to develop these twins in healthcare and assess their credibility. Hence, it should entail the development of a federated public infrastructure and the collection of said resources, driven by the engagement of a collaborative ecosystem.
In order to realise the potential of the VHT, a shared vision and inclusive eco-system driven roadmap has been developed, supported through the EDITH Coordination and Support Action. The roadmap contains a blueprint of the Virtual Human Twin and will identify the required (technical) developments, including but not limited to interoperability, computability, hardware and integration of health models & data. Ethical, Legal and Social elements are discussed, including privacy, intellectual property rights, standards, regulations, ethics and social acceptance. The user perspective is developed for a range of stakeholders, whose needs and value propositions have been identified through a range of activities and interactions. Finally, sustainability of the ecosystem and infrastructure are examined, looking at incorporation in clinical and industrial workflow, development of economic activities, a member-state strategy and the creation of a public research infrastructure organisation. Recommendations for all involved stakeholders have been formulated. The final draft of the roadmap can be found here: https://zenodo.org/records/14645647.
In this talk I will discuss the current status and challenges that lie ahead on the road to realization of the vision of the Virtual Human Twin, using examples of my own research group as well as the work of colleagues.
Liesbet Geris
University of Liège, KU Leuven & VPH Institute, Belgium
Tomasz Gosiewski – Jagiellonian University Medical College
The digestive tract microbiome – where does it come from, how does it change, and what is its connection with the brain?
Microbiome it is a mysterious phenomenon that is talked about and written about in the media. Often mistakenly referred to as flora or microflora, but it has nothing to do with plants. So what is the microbiome? It is a large collection of genes of microorganisms that inhabit their host and usually do not harm it, but on the contrary, cooperate with it. Sometimes another term also appears, namely microbiota. The microbiota is nothing more than all the cells or viruses that inhabit their host. So why two separate terms, the microbiome and the microbiota? Because by using research methods based on genetic analysis, we study the genes of microorganisms, i.e. the microbiome, and by detecting individual cells of microorganisms directly, we describe the microbiota. However, genetic studies provide a more comprehensive understanding of our microscopic neighbours, so they are preferred by scientists.
We acquire the microbiome at birth, because this is when the newborn has “first contact” with microorganisms, although some believe that this happens while still in the womb. What this contact will be like depends on the type of birth, and the optimal one is the natural one. As it passes through the birth canal, the newborn and its digestive tract are colonized by bacteria that inhabit the vagina, which are the beginning of the normal microbiome. If the mother feeds the newborn with her own milk, she additionally supports the developing microbiome. Delivery by caesarean section causes bacteria from the hospital environment (pathogenic, multidrug-resistant) to enter the newborn’s digestive tract first and they initiate the microbiome (abnormal). The microbiome profile in the first days and weeks of life determines the likelihood of developing inflammatory diseases later in life. An abnormal gut microbiome increases the risk of diseases such as diabetes, obesity, inflammatory bowel disease, allergies, autoimmune diseases, autism, and many others.
Is the microbiome related to the brain? It is surprising, but scientific discoveries indicate that it is. The composition of the gut microbiota influences the dense network of neuronal connections that wraps around the gut. Neurons produce neurotransmitters such as serotonin, norepinephrine, dopamine and others. The microbiota influences the chemical profiles of neurotransmitters in the gut, which in turn transported through the blood to the brain and affect it. On the other hand, the brain, through the vagus nerve, influences intestinal peristalsis and, therefore, the microbiota. These interactions are being studied intensively and are an exciting field for neuroscience and neuropsychoatrics.
Tomasz Gosiewski
Head of Microbiome Research Laboratory; Chair of Microbiology; Molecular Medical Microbiology Department
Faculty of Medicine Jagiellonian University Medical College
Alfons Hoekstra – University of Amsterdam
Towards Digital Twins for Cerebral Blood Flow and Perfusion Pathologies
A multiscale computational model for for cerebral blood flow, perfusion, and tissue metabolism will be introduced. The main components are: (1) a 0D blood flow model including the heart, large systemic arteries, the Circle of Willis, and smaller cerebral arteries projecting on the pial surface; (2) a 3D brain perfusion model using a three-compartmental porous medium approach capturing the length scales of the arterial, capillary, and venule vessels of the brain; and (3) a tissue metabolism and tissue death model. Details of the components and their couplings will be discussed, as wel as validation results. Two examples of digital twin using these computational models will be introduced: (1) estimation of infarcts in acute ischemic stroke patients (using the fully coupled model); (2) estimation of orthostatic hypotension in elderly individuals (only relying on the 0D blood flow component).
Joanna Jaworek-Korjakowska – AGH University
Artificial Intelligence in Orthodontics: From Automated Diagnostics to Personalized Treatment
Artificial intelligence (AI) is increasingly transforming orthodontics by enabling more precise diagnostics, treatment planning, and patient monitoring. Machine learning algorithms applied to radiographs, 3D scans, and intraoral images can automatically detect anatomical landmarks, classify malocclusions, and predict treatment outcomes with high accuracy. Deep learning models facilitate automated cephalometric analysis, reducing human error and saving clinical time. In addition, AI-powered simulation tools allow for individualized treatment planning, such as predicting tooth movement and optimizing aligner design. Beyond diagnostics and planning, AI supports remote monitoring through image-based progress tracking, enhancing patient engagement and enabling timely interventions. Ethical and clinical considerations, including data privacy, bias, and explainability, remain central to integrating AI safely into practice. Overall, AI offers significant potential to improve efficiency, accuracy, and personalization in orthodontic care, paving the way for more accessible and patient-centered treatments.
Emiliano Ricciardi – IMT School for Advanced Studies Lucca, Lucca, Italy
Modeling the Sensory-Deprived Brain: Insights from Neuroimaging, Computational Neuroscience, and Machine Learning
How does the brain adapt to the absence of an entire sensory modality from birth? This question has long fascinated neuroscientists interested in the balance between neural plasticity and biological constraints. In this talk, I will present recent efforts from our group at the IMT School for Advanced Studies Lucca, where we integrate high-resolution neuroimaging and machine learning approaches to explore how the brain reorganizes itself under conditions of congenital sensory deprivation.
Combining techniques such as multivariate pattern analysis, machine learning approaches and functional network modeling to neuroimaging data, we describe how distributed brain networks reorganize and how conceptual knowledge emerges independently of visual input. We then feed these neural signals into deep generative models, allowing us to reconstruct high-level visual representations from brain activity in congenitally blind individuals.
These interdisciplinary approaches reveal a dual nature of cortical organization: a stable, hierarchically structured architecture that persists without sensory input, and a flexible layer of experience-driven reallocation. By bridging empirical and computational neuroscience, our work contributes to a deeper understanding of how the human brain maintains functional integrity and cognitive richness even in the face of profound sensory absence.
Daniel Taylor – University of Sheffield
Computing coronary physiology: Conception, Optimisation and Clinical application
Computed coronary physiology (virtual FFR) is now established in clinical practice. Our group recently developed a model for predicting absolute flow and hyperaemic coronary microvascular resistance using pressure wire and angiographic data. In this talk I will first outline how we developed the model and implemented side branch flow. I will then cover the optimisation and validation of these advancements. Finally, I will discuss how the model may be used to enhance clinical practice, by applying it retrospectively to data from the landmark ORBITA trial. Specifically, I will explore the relationship between disordered coronary microvasculature and the symptomatic response to percutaneous coronary intervention in patients with stable angina.
Honorary patronage


The conference is held under the honorary patronage of the Mayor of the City of Kraków, Aleksander Miszalski.
Media patronage

No registration fee, conference by invitations
Regulations Krakow Conference on Computational Medicine 2025


